Professor Gary Carvalho
Emeritus Professor

Affiliations
Contact info
Position: Emeritus Professor of Molecular Ecology
Email: g.r.carvalho@bangor.ac.uk
Location: ECW 3rd floor
Phone: +44(0)1248 382100
Web: Personal Web Page
Contact Info
Position: Emeritus Professor of Molecular Ecology
Email: g.r.carvalho@bangor.ac.uk
Location: ECW 3rd floor
Phone: +44(0)1248 382100
Web: Personal Web Page
Overview
Prof. Gary Carvalho undertakes research on the molecular genetic analysis of population and species biodiversity of aquatic animals, with studies aimed at understanding the evolutionary and ecological forces that shape genetic structure in the wild, and how such structure may influence adaptation, population persistence and distribution.
The cornerstone of Professor Carvalho’s research has focused on the dynamics and significance of genetic diversity in wild aquatic animals using molecular markers. Effort is placed on demonstrating the value of molecular tools and genetic diversity as core components of environmental management and conservation of biodiversity, especially among commercially exploited species. Moreover, throughout his career, he has engaged with governance and policy making to foster awareness and the inclusion of genetics in environmental science. The underpinning strategy has been to elucidate drivers of genetic change, especially at the population level, and then to apply such understanding in the conservation of genetic resources. Co-Editorship of Special Issues at the interface between science and policy (e.g. 1995, 2008, 2011, 2015, 2016) have aimed at making genetic concepts and tools more accessible to marine wildlife, fisheries and aquaculture managers, environmental and conservation biologists. Two key messages encapsulate his research: first, demonstration of the speed at which genetic change can occur, and second, the association between biological traits and the dynamics of genetic diversity, especially in changing environments. Insights have been generated by state-of-the-art methods, (e.g. genetic fingerprinting; transcriptomics; 454 sequencing; resting egg banks; ancient DNA (aDNA) retrieval; gene-associated SNPs; environmental DNA (eDNA)), often in the frame of pioneering studies, to unravel the dynamics of genes in experimental and natural settings.
Further details are available under my public profile and CV.
About my research
Using primarily DNA-based tools, my research is aimed at the elucidation of fundamental aspects of a species’ biology such as patterns of dispersal and gene flow, evolution of life histories and behaviour, response to environmental stress, and mechanisms of speciation, as well as the application of molecular tools to the management and conservation of exploited aquatic species from temperate, tropical and Antarctic marine and freshwater ecosystems.
Research includes the molecular analysis of population and species biodiversity of aquatic animals, with studies aimed at understanding the forces that shape genetic structure in the wild, and how such structure may influence adaptation, population persistence and distribution. Notable areas of activity include: the evolution and ecological significance of population differentiation, phylogeography and phylogenetics of aquatic taxa, the molecular analysis of past populations using PCR-based recovery of DNA (ancient DNA) from resting eggs and preserved material (e.g. fish otoliths and scales), DNA barcoding, traceability of fish and fish products, the evolutionary genetics of clonal animals, the evolution of adaptive traits using molecular, genomic and quantitative genetic analysis, and fisheries and conservation genetics of exploited fish in temperate, tropical and Antarctic waters.
Carvalho led an EU consortium FishPopTrace, (2008-2011) with among the first applications of single nucleotide polymorphisms (SNPs), providing new approaches to tackling structuring in the marine environment and the identification of candidate genes.
Professor Carvalho continues as an Editor of the Proceedings of the Royal Society London, B (PRSB), Editor of the PRSB Evidence Synthesis articles, and Editor of Fish and Fisheries. He is currently the Honorary Past President of the Fisheries Society of the British Isles, and is a member of the Scientific Steering Committee of the World Fisheries Congress to be held in Seattle, in March 2024.
If you want to find out more, click on the research tab above.
A past significant international event coordinated by the former Molecular Ecology and FIsheries Genetics Laboratory (now, MEEB), MEFGL was a major international symposium on Fish, Genes and Genomes in July 2016 with leading key-note speakers such as Axel Meyer, Robin Waples, Craig Primmer, Louis Bernatchez, Jenny Ovenden and John Casey.
Other
Research
Much of what is detailed below documents major past contributions of the former, Molecular Ecology and Fisheries Genetics Laboratory (MEFGL), now the Molecular Ecology and Evolution@Bangor (MEEB) research group. For current activities please visit the MEEB homepage.
I arrived in the former School of Biological Sciences, University of Wales Bangor, in January 2005, having moved from the Molecular and Evolutionary Ecology Group at Hull University. My initial remit was to expand in particular, studies on the molecular ecology of aquatic animals, including the use of molecular markers in fish and fisheries genetics. Although most research projects incorporate DNA-based tools, my research is aimed at the elucidation of fundamental aspects of a species’ biology such as patterns of dispersal and gene flow, evolution of life histories and behaviour, response to environmental stress, and mechanisms of speciation, as well as the application of molecular tools to the management and conservation of exploited aquatic species from temperate, tropical and Antarctic marine and freshwater ecosystems. In addition to existing areas of interest, the position at Bangor provided additional opportunities to develop studies on functional aspects of adaptive variation through genomic analysis, links between dispersal, gene flow and physical oceanography in the sea, and the use of high throughout sequencing.
The Bangor-based Molecular Ecology and Evolution@ Bangor research group (MEEB) now represents among the largest European groupings, which in recent years has expanded beyond the former MEFGL-focus on aquatic taxa to encompass microbial communities, including those associated with disease, reptiles and plants.
The purpose-built suite of molecular ecology laboratories in the Environment Centre Wales (ECW) provides a fully integrated and dedicated platform for the molecular analysis of biodiversity in wild populations, linked to other university genomic facilities. In addition to on-going work on fundamental evolutionary processes more opportunities are available for engagement with environmental agencies through links with the Centre for Ecology and Hydrology (NERC), Environment Agency (UK), Forestry Commission and Natural Resources Wales (NRW), all of which have representation within ECW. Additionally, we have string and productive links with UKRI and other genomic facilities, providing access to training and facilities for state of the art genomic analysis and bioinformatics.
Below I summarise some ongoing areas of interest:
Molecular analysis of population structure

Molecular markers can be used to examine the relative roles of various microevolutionary forces on the levels and significance of genetic differentiation among populations. For example, one past project, in conjunction with the British Antarctic Survey and University of Hull, aims to disentangle the relative roles of life history variation and hydrographic processes on dispersal and gene flow in Antarctic fishes. It is becoming increasingly apparent that despite high dispersal capacity, some marine species continue to exhibit significant population structuring, occasionally on fine geographic scales. We also examined, in collaboration with the University of Aveiro, links between hydrographic variability and larval dispersal and recruitment in the shore crab Carcinus maenas. In addition to exploring the role of hydrography in dispersal, we have assessed methods for population assignment, and in conjunction with fine-scale genotypic monitoring and the role of selective mortality in the early stages of population recruitment.
Use of ancient DNA to study long-term environmental change
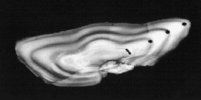
The availability of PCR enables recovery of DNA from natural (e.g. resting eggs of zooplankton: e.g. rotifers) and archived (e.g. fish scales and otoliths: e.g. cod) material to examine long-term changes in genetic structure. Using such approaches it is possible to examine the impact of natural and man-made environmental change on population and community structure. In collaboration with CEFAS we developed robust and sensitive methods for recovering DNA from archived histological samples of the flatfish, dab, Limanda limanda, as a basis for comparing genetic structuring in contemporary and past populations.
Evolution of adaptive variation

A core approach in molecular ecology has been to utilise neutral molecular markers to generate inferences about adaptive variation. Other approaches include either direct analysis of gene function or the use of quantitative genetic approaches to identify quantitative trait loci or to estimate heritabilities in biological traits. In conjunction with colleagues at the University of Hull we have for example: (1) examined geographic variation and performance under semi-controlled conditions of a key gene (Pantophysin I) associated with growth rate variation, (2) examined adaptive variation in freshwater bullhead using a combination of neutral and quantitative traits to assess heritabilities in relation to habitat variation.
We developed among the first studies to employ digital transcriptomics to explore the impact of environmental stress on gene expression and associated adaptive phenotypes. Past systems include Nucella lapillus and differential gene expression in response to environmental changes (natural and man-made), including the impact of contaminants on sex reversal (“imposex”), and impact of environmental stress on gene expression in marine nematodes. We were among the first to generate a transcriptome of a marine gastropod, and in early 2013, one of the major outcomes of the work was published in Molecular Ecology, alongside a Perspective to explore its wider contribution to the field of genomics in non-model species. The paper: “Transcriptomics and in vivo tests reveal novel mechanisms underlying endocrine disruption in an ecological sentinel, Nucella lapillus”, provides novel insights into the control of imposex, and importantly, yields evidence for a common signalling pathway between invertebrate and vertebrate species that has previously been overlooked in the study of endocrine disruption. With funding from the NERC, and in conjunction with the former Liverpool Microarray Facility, we developed microarrays to explore genetic changes associated with phenotypic shifts associated with anadromy in brown trout, Salmo trutta.
Fisheries Genetics and Conservation
We have utilised a plethora of molecular genetic markers to examine the stock structure of commercial fishes, as well as examining demographic processes that may affect response to harvesting and environmental change. In particular we have utilised archived scales and otiliths to explore changes in genetic diversity and structure in relation to over-exploitation, and have demonstrated not only rapid short-term genetic change in North Sea cod during periods of population decline, but also a loss of genetic diversity following population collapse in New Zealand snapper. In conjunction with the Environment Agency, UK, our work extended to examining population structure and biodiversity of salmonid fishes.

Many global marine fisheries have collapsed, or are at record low levels of abundance. Continuing exploitation and the uncertain impacts of climate change are adding further pressures on fish resources. New strategies are therefore required to assist in our management and conservation efforts. One such development will be to incorporate the extent and dynamics of spatially-associated biological differences that exist among fish stocks into stock assessment. Such information is important since most exploited fish species comprise assemblages of individuals that differ in their vital rates of growth, reproduction, migratory tendency and mortality. It therefore becomes desirable, for example, to match the level of fishing intensity to the projected rate of replenishment following harvesting, thereby reducing the probability that individual stocks will become extinct. Conserving such biological differences among stocks is also important in the maintenance of genetic diversity in wild fish populations, so endowing them with greater evolutionary potential for adapting to changes in the environment. A critical component of such work involves the testing of hypotheses relating to the relative contributions of such factors as dispersal of eggs and larvae, and behaviour of juveniles and adults in maintaining such biological differences. By integrating research efforts across each of the three major UK fisheries agencies (FRS, CEFAS, AFBI) with partners in UK Universities, a NERC Institute and an international cod expert from Denmark, we examined for the first time the extent, patterns and stability of cod (Gadus morhua) population structuring throughout UK waters.
The proposal represented among the first opportunities for UK Universities and all three UK Fisheries agencies to work together on a problem that has to take account of the interdependence of fish stocks across large regions of UK and adjoining waters. The proposal generated new genetic estimates of stock separation, especially in areas of uncertainty such as the Celtic and Irish Seas and the Southern North Sea and English Channel, as well as the provision of new theoretical tools (population models) than can be used to forecast the impact of continued fishing pressure and environmental change on cod populations. Moreover, the tools provided information on the appropriate spatial scale and distribution of marine protected areas and the probable rates of stock recovery in a species that is now formally endangered and listed in the IUCN Red List.
A past EU FP7 project, AquaTrace, in partnership with a large consortium of European workers who aim to develop novel tools to explore the impact of aquaculture on native marine fish populations. The Bangor-UEA component of the work focused on the phenotypic and genetic basis of fitness shifts associated with introgression between wild and captive-bred Atlantic salmon. The latter work was carried out in conjunction with colleagues at the Bergen Marine Labaoratory, and will utilise state-of-the-art common garden facilities to explore fitness consequences and to map quantitative trait loci.

To further illustrate some past work on adaptive diversity in the wild, a fomer PhD project examined the genetic basis of fisheries-induced evolution, using the Trinidadian guppy, Poecilia reticulata, as a model system. Size-selective harvesting can induce rapid phenotypic changes, such as age and size of maturation, in exploited fish populations. Despite the increasing incidence of such shifts, the relative contributions of genetic and environmental factors remain uncertain and are much-debated. Outputs from the study will appear early in 2013 in the journal, Frontiers in Ecology and the Environment, and demonstrate a significant genetic component to fisheries-induced evolution in an experimental system, with major implications for the sustainability of exploited populations, as well as impacts on size-structured communities and ecosystem processes, prompting a reconsideration of adaptation to, and recovery from, harvesting and predation. The work was extended to another PhD studentship which examined in more detail the genomic shifts associated with fisheries induced evolution in the model guppy system.
Traceability of fish and fisheries forensics

The former MEFGL was awarded (2008–2011) a major EU grant (FishPopTrace) to examine the traceability of fish and fish products in European waters, with focus on herring, cod, hake and sole. The 15 partner consortium was coordinated by Gary Carvalho and the former MEFGL, developed for the first time a forensic framework for legal enforcement of policies aimed at reducing the amount of Illegal, Unreported and unregulated fishing (IUU). There is an increasing requirement for traceability of fish and fish products, both for consumer protection and for regulatory enforcement. For example, in the UK, the Marine Stewardship Council encourages consumers to eat particular landings of cod that are taken from ‘stocks maintained within safe limits’. A traceability system based on regional stocks is necessary to preclude fraudulent allocations. The underlying rationale of FishPopTrace was to assess and address challenges arising from the development of traceability tools within a forensic framework for four judiciously chosen target species: cod (Gadus morhua), hake (Merluccius merluccius), herring (Clupea harengus) and sole (Solea solea). While current information on levels of population structuring in traits such as life histories, morphometrics, genetics and physiology were used to inform sample choice, new data was restricted to markers at two levels: (1) Routine screening: selection of markers that exhibited maximal discriminatory power to identify populations, though with discrete and controlled variance enabling validation (single nucleotide polymorphisms (SNPs) and otolith microchemistry and morphometrics). Data from DNA-based methods provide a mechanism for traceability throughout the food supply chain (“fish to fork”) and indicate discrete spawning populations, whereas otoliths provide an independent on-board traceability system of fish provenance. (2) Testing of novel tools: additional tools were tested on a selection of populations to assess validity and potential for traceability and validation, including fatty acid analysis, proteomics, gene expression analysis and the generation of high-throughput microarray platforms for SNP genotyping. FishPopTrace yielded novel information that relates to geography (“population tag”), as well as providing regional signatures that indicated biological differentiation in relation to spawning identity. Specifically, the consortium brought together past expertise in fish traceability projects (Fish and Chips (GOCE-CT-2003–505491), FishTrace (QLRI-CT-2002–02755, FISH-BOL, Consortium for the Barcode of Life) to address several inter-related objectives:
1.1.2.1 To integrate recent and on-going data from European fish species traceability projects, and to generate a single compatible data base and tissue archive managed by the Joint Research Centre of the European Commission. The outputs will comprise a new data base and associated web links with access to recently generated data on fish species and population identity, together with an archive of associated tissue samples from external and consortium outputs.
1.1.2.2 To examine single nucleotide polymorphisms (SNPs) and otolith microchemistry and morphometrics in widely distributed populations of cod, hake, sole and herring, as tools for discriminating biologically differentiated populations and as a basis for traceability. Outputs will comprise population-level signatures associated with fish origins in early life and representative spawning groups.
1.1.2.3 To undertake validation of traceability tools in relation to end-user technology. Outputs will produce Standard Operating Procedures (SOPs) to allow transfer of technologies to other laboratories throughout EU member states.
1.1.2.4 To develop a population monitoring system based on otolith and genetic data that will assess population stability in a temporal and spatial framework. For each species, alternative parameters will be identified as indicators of population stability, and parameters will be validated using a combination of archived data and tissue samples.
1.1.2.5 To test the utility of additional novel traceability systems (fatty acid profiles, proteomics, gene expression, microarray platform for SNP genotyping). Outputs will comprise an assessment of the utility of additional novel approaches to traceability and population monitoring through estimating the within and among-population components of biological differentiation and population signatures. Additionally, a chip-based platform will be generated to undertake high-throughput SNP genotyping.
1.1.2.6 To facilitate technology transfer in relation to enforcement and conservation policies of the CFP and associated socio-economic consequences. Central elements of the output will be the development and evaluation of end-user tools, a Cost Benefit Analysis and a final report setting FishPopTrace in the context of the CFP.
A major output from the project was published in 2102 in Nature Communications. By applying high differentiation single nucleotide polymorphism assays, in four commercial marine fish, on a pan-European scale, we find 93–100% of individuals could be correctly assigned to origin in policy-driven case studies. We showed how case-targeted single nucleotide polymorphism assays can be created and forensically validated, using a centrally maintained and publicly available database. Our results demonstrated how application of gene-associated markers will likely revolutionize origin assignment and become highly valuable tools for fighting illegal fishing and mislabelling worldwide.
Molecular Identification, DNA Barcoding and Phylochips

The use of short, standardised short DNA sequences – a “DNA barcode” has become a valuable addition to the taxonomic toolbox to validate species identity, often these days extended include metabarcoding, as well as to identify unknown species, cryptic life history stages, etc. The Consortium for the Barcode of Life (CBOL) launched a new initiative in 2005 to DNA barcode all fishes- “FISH-BOL”, which will generate a global database for biologists and interested naturalists. In the former MEFGL, I had the pleasure of acting as founder Chair of the European Regional working group for FishBol between 2005–2011. Our DNA barcoding of marine fish was expanded to include novel data on a hot-spot of biodiversity – the Indo-Malay Archipelago. Some of the findings from a past PhD programme with a local Malaysian student based in the former MEFGL, was published in 2012 in PLoS ONE, and represents among the first comprehensive analyses of cryptic species in a large commercially important group, the Carangidae. Other DNA barcoding outputs included the Crustacea, esp. Decapoda, from the completed 2012 PhD thesis of a former MEFGL-Portuguese FCT student, generating several papers detailed in the Carvalho publication list.
Personal
Education and honours
- Fellowship of the Linnean Society (F.L.S.) (2000)
- Fellowship of the Society of Biology (F.S.B.) (2011)
- PhD. (Wales, 1985) – Ecological genetics of Daphnia
- M.Sc. (Wales, 1980) – Ecology
- B.Sc. (Class I, London, 1978) – Environmental Biology
Career
- 2009– Visiting Professor, University of Penang, Malaysia
- 2005–present Professor of Molecular Ecology, School of Biological Sciences/Environment Centre Wales, Bangor University
- 2006–2008 Deputy Head of School of Biological Sciences
- 2007- Chair, Environment Centre Wales, Management Board
- 1996–2004 Professor of Molecular Ecology, Dept. Biological Sciences, Hull University
- 1998–2004 Research Director, Department of Biological Sciences, Hull
- 1995–1996 Senior Lecturer in Marine Biology, University of Wales, Swansea
- 1990–1995 Lecturer in Marine Biology, Biological Sciences, University of Wales, Swansea
- 1988–1989 AFRC R.A. (University of Southampton), DNA fingerprinting of aphids
- 1986–1988 NERC Post-doctoral Fellowship (Univ. Wales, Bangor), population structure of isopods
- 1986–1987 Max-Planck-Society Research Fellowship (M.P.I. of Limnology, Germany), Clonal Structure of Daphnia
Editorial Work
- 1995–2000 Asst. Editor (Genetics and Evolution): Journal of Fish Biology
- 1996–2000 Editorial Board: Reviews in Fish Biology and Fisheries
- 1998–2003 Editorial Board: Heredity
- 2000 -present Editorial Board: Molecular Ecology
- 2000–2007 Associate Editor: Fish and Fisheries
- 2000 – present Editorial Board: Conservation Genetics
- 2003–2009 Editorial Board, Proceedings of Royal Society, London, B.
- 2007- present Editor, Fish and Fisheries
- 2011- present Editor, Proceedings of Royal Society, London, B.
- 2012–2013 Editorial Board, Biological Invasions
External Professional Activities
- 1998-present ICES Working Group on the Application of Genetics in Fisheries and Mariculture
- 2000–2007 NERC Steering Committee: Environmental Genomics Thematic Prog.
- 2003- NERC ad hoc Advisory Committee on Post-genomic Science and Proteomics
- 2003–2007 Management Group of Directors Working Group (CEFAS, DEFRA, DARDNI, FRS) on Cod Population Structure
- 2004–2008 Scientific Advisory Panel, Environment Agency
- 2004–2007 NERC Steering Committee, UK Molecular Genetics Facilities
- 2005–2009 Chair, European Coordinator of FISH-BOL (DNA Barcoding of Marine Fishes) and Steering Committee member
- 2005–2008 NERC Peer Review College
- 2005–2006 Vice-President of the Fisheries Society of the British Isles
- 2005–2006 Member of SEBI2010 Expert Group on Fishes (EEA/ECNC/UNEP). Streamlining European 2010 Biodiversity Indicators.
- 2006- Expert Evaluator Panel- Norwegian Research Council (Seas and the Oceans)
- 2007- Member Academy of Finland Biosiences Grant Review Panel- Ecology Panel
- 2007- Chair of NERC Moderating Panel A (Molecular Ecology & Evolution)
- 2008-present External Consultant on various programmes for FCT (Portuguese Research Council for Science and Technology)
- 2008- ERA-Net BiodivERsA Evaluation Committee (EU Commission)
- 2008–2011 Coordinator of EU FP 7 Consortium, FishPopTrace
- 2008: Member of Research Quality Review, University College Cork
- 2009–2011 FAO Expert Group on Fisheries Forensics
- 2009 Visiting Professor, University of Penang, Malaysia
- 2009–2010 Expert Group of Marine Biodiversity for IFREMER, French Ministry of the Environment
- 2010 Organising Committee,SNP III International Conference Seattle, Washington
- 2010 Organising Committee, ECBOL2:2nd European Conference on DNA Barcoding
- 2000-present Editorial Board: Molecular Ecology
- 2000–2007 Associate Editor: Fish and Fisheries
- 2008-present Editor: Fish and Fisheries
- 2000-present Editorial Board: Conservation Genetics
- 2003–2009 Editorial Board, Proceedings of Royal Society, London, B
- 2011-present Editor, Proceedings of Royal Society, London, B
- 2011 Guest Editor, Applications of SNP Genotyping in Non-model organisms, Molecular Ecology Resources
- 2012 Editorial Board, Biological Invasions
- 2012 – Founder Member – UK Biodiversity Science Committee – to represent the UK in the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystems (IPBES) and other International biodiversity initiatives
- 2012 – European Expert Group on Fish Stock Assessment (Joint Research Centre- “Assessment for All”- EC)
- 2012 – DEFRA Stakeholder Advisory Group on Fish Traceability
- 2012 – Appointment by Centre for Independent Experts, US – scientific advice on petition for two species of river herring (blueback and alewife) to be included under Endangered Species Act
- 2012–2014 – NERC Peer Review College and Panel Member, Natural Environment Research Council, UK
- 2013 – FAO Expert Advisor on Establishment of Advisory Group on Aquatic Genetic Resources, Bangkok
- 2013, 2014 – Chair, Academy of Finland Biosiences Grant Review Panel- Ecology Panel
- 2013 – Member of the Advisory Board and Task Force for the Natural Environment Research Council (NERC, UK) knowledge exchange program on sustainable fisheries & aquaculture
- 2013 – Co-Convenor of Congress of the European Society for Evolutionary Biology, special symposium: Genomic Islands: their role in adaptation and speciation (August, Lisbon)
- 2013 – Member and Chair of Review Panel, Belgian Research Action through Interdisciplinary Networks, Belgian Government, Belgian Science Policy Office, Brussels
- 2013, 2014 – Member of the European Commission Evaluation Panel for Marie-Curie Actions, Environment. Brussels
- 2013 – Independent External Advisor, Marine Stewardship Council, DNA testing and certification of sustainable fisheries
- 2014 – present – NERC Core Panel Member – Panel E and others.
- 2014–2017 – Chair, ICES Working Group on the Application of Genetics in Fisheries and Mariculture (WGAGFM)
- 2014 – Founder Member of Species Survival Commission (IUCN) Specialist Group in Conservation Genetics.
- 2015 - OECD Expert Group Member on the long-term potential of marine biotechnology
- 2015-2018 - Vice-President, Fisheries Society of the British Isles
- 2016 - Co-Convenor Bioeconomics, Sociobiology and Other Mixes.The Advantage of Linking Disparate Data to Gain New Insights into the Exploitation of Marine Fish. Special Theme Session, World Fisheries Congress, South Korea (May, 2016)
- 2016 – Convener, Fisheries Society of the British Isles (FSBI) Annual International Symposium: Fish, Genes and Genomes: Contributions to Ecology, Evolution and Management. Bangor, UK, July 2016.
Outreach and Public Communication of Science
a. Communicating genetic principles to environmental managers
For over 2 decades, effort has been focused on communicating the value of molecular tools and genetic diversity as core components of environmental management and conservation of biodiversity, especially among exploited species. Targeted publications such as Special Issues of international fisheries journals (e.g. 1995, 2008) aimed to render genetic concepts and tools more accessible to fisheries managers and conservation biologists. Direct input has been facilitated by engagement in appropriate bodies charged with responsibility for managing natural resources (e.g. ICES Working Group on the Application of Genetics in Fisheries and Mariculture; Management Group of Directors Working Group (CEFAS, DEFRA, DARDNI, FRS) on Cod Population Structure; Member of SEBI2010 Expert Group on Fishes (EEA/ECNC/UNEP). Streamlining European 2010 Biodiversity Indicators; FAO Expert Group on Fisheries Forensics; Expert Group on Marine Biodiversity for IFREMER, French Ministry of the Environment, 2009–2010; Founder Member – UK Biodiversity Science Committee – to represent the UK in the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystems (IPBES) and other International biodiversity initiatives; Expert Food and Agriculture Organization of the United Nations State Of World Consultation on Aquatic Genetic Resources, Bangkok).
In addition to various research projects focusing on fisheries genetics, a notable recent programme, FishPopTrace, provided an opportunity for directly influencing the revision of major policies (Revised EC Common Fisheries Policy). FishPopTrace, an EU FP7 project (2002–11), coordinated by Carvalho, aimed to generate forensic tools (primarily single nucleotide polymorphisms, SNPs), to deliver a breakthrough in the legal enforcement of regulatory policies within the fishing industry. The key quest was to trace fish back to localities or spawning populations: the target and unit of harvesting and spatially resolved policies. For the first time, by applying high differentiation SNP assays, in four commercial marine fish, on a pan-European scale, it was possible to correctly assign unknown individuals back to their source spawning populations with unprecedented accuracy (93–100%). Outputs demonstrated how application of gene-associated markers will likely revolutionize origin assignment and become highly valuable tools for fighting illegal fishing and mislabelling worldwide. Recognising the impact of FishPopTrace (e.g. in Annex 1of the Commission Regulation for establishing a control system to secure compliance with the rules of the Common Fisheries Policy (COM (2008)721 final), it was stated “It is interesting to note the innovative contribution of FishPopTrace …for implementation of modern technologies”. Such distinction, combined with the new regulations imposed by the EC (EC Regulation 1224/2009 (Art 13), requiring EU States to undertake pilot studies of novel traceability tools by 2013, the Department of Environment Food and Rural Affairs (DEFRA, UK Govt.), is tailoring FishPopTrace tools for use by the UK fishing industry. FishPopTrace, and direct partnership with the Joint Research Centre (the science wing of the European Commission), also provided opportunities for direct input to consultation exercises on the Revised Common Fisheries Policy, in particular the successful adoption of advanced technologies for use in traceability and legal enforcement of regulations covering importation and capture of fish and fish products. The FishPopTrace SNP tools are also now being extended to new marine fish species around Europe (sea bream, bass, turbot), to generate a DNA reference data base for tests of traceability of escapee fish back to fish farms, as well as monitoring impact on fitness of wild fish species.
In July 2013, Carvalho was appointed by the Marine Stewardship Council as an Independent External Advisor on DNA testing of fish and fish products in the context of certification of sustainable stocks. Recent public and Government concern over traceability of food products, in conjunction with sustainability targets, demands a concerted, transparent and robust application of forensic standard authentic tests.
b. Broader outreach activities
Societal interest in biodiversity and management of natural resources has secured regular opportunities for communicating scientific outputs to the general public directly and through the media. Most notably, since 2010, contributions have been made to the week-long Bangor Science Festival, through evening public lectures on topics such as sustainability and fisheries forensics, as well as engagement with school children to introduce them to the excitement and wonder of the natural world through illustrating the uses of DNA tools to environmental management. The recent award of a Royal Society Partnership Grant, for partnership with a comprehensive school on Anglesey (the first such award in North Wales), has provided an interactive opportunity to discuss our research on “climate change, Antarctica and ice fish” – all topics that generate curiosity and interest, especially how scientific evidence can be gathered to support notions of climate change. In 2013, we coordinated a day-long event (with over 500 public visitors), as part of an event “The Hidden World”, where we provided hands-on and interactive displays illustrating life in the Antarctic, the life history of ice fish, and how climate change will likely impact these fragile marine ecosystems. Pupils aged 13–15 from our local partnered School presented talks to the general public on their own work to date as part of the Partnership Grant.
There has also been much media interest in our work– including interviews on BBC Radio on advances in using genetic fingerprinting to monitor the invasion of pest aphids (BBC Radio Wales; various national newspapers); the use of allozymes to manage and conserve the Falkland Island Fishery (BBC North East, local news); and more recently the generation of DNA-based tools for use in tackling illegal fishing and eco-certification of fish and fish products: (BBC World Service; Science In Action: http://www.bbc.co.uk/programmes/p00s1t04); Radio 4, BBC Farming Today; episode of the BBC Science Programme, in May 2013, Bang Goes the Theory, on the use of DNA forensics in traceability of fish products; NERC podcast on illegal fishing; http://planetearth.nerc.ac.uk/multimedia/story.aspx?id=452), as well an numerous regional and national newspapers.
Research outputs (147)
- Published
An integrated spatio-temporal view of riverine biodiversity using environmental DNA metabarcoding
Research output: Contribution to journal › Article › peer-review
- Published
Nanopore environmental DNA sequencing of catch water for estimating species composition in demersal bottom trawl fisheries
Research output: Contribution to journal › Article › peer-review
- E-pub ahead of print
Comparative population genomics of manta rays has global implications for management
Research output: Contribution to journal › Article › peer-review
Projects (17)
KESS II PhD with Dwr Cymru BUK299
Project: Research
CASE studentship award - William Perry
Project: Research
Genetic tools to resolve taxonomy of Manta and Mobula rays
Project: Research
Media coverage (1)
Great auk extinction: Humans wiped out giant seabird
Press/Media: Research

